This lab presumes an understanding of certain basic concepts, which were covered in the lecture material associated with this module:
- Mean Information Field
- Diffusion
- Contagious Diffusion
- Hierarchical Diffusion
- Diffusion waves
- Social Gravity model
- Simulation
- Geographic Distribution of Population
- Urban Hierarchy (e.g. Central Place)
- Rank/size distribution
- Logistic curve
Lab Objectives:
In this lab, you will
- Run the demo of the Diffuse software, for a simulation of a cholera epidemic.
- Run Diffuse on cholera, changing parameters (especially the hierarchical and distance-decay parameters) and remarking on the subsequent changes in the results. You will compare your results with others via a web-based form.
- Compare Moran's I computed using Hierarchical and Contagious Diffusion contiguity matrices, using Stat!, to determine if we can tease out the components in a model you will simulated.
Lab
- Download the Diffuse software to your machine.
UMich Students: save it to c:\temp.
Run the demo provided, of cholera. Take all the defaults, don't intervene, etc.
- Now run Diffuse again
- Decline to execute a demonstration (n).
- Study cholera (y).
- Don't input your own population map (n).
- Choose the clustered pattern distribution (2).
- Choose a saturation proportion: (.8).
- Use a distance-decay diffusion model (2).
- Choose distance decay parameter between .1 and 1.5 (make your own choice, and note it).
- Choose hierarchy parameter between 1.0 and 3.25 (make your own choice, and note it).
- We'll choose the co-ordinates of the infected person (10,4 - don't use parentheses! Just separate them by a comma....).
- No quarantine! (n)
- No intervention! (n)
- Hit return until you see the S-shaped
(logistic) curve of cumulative cases: from this you can see what some
interesting points in the progression of the disease (the point at which it
took off, the point at which it began to level off, etc.). Make a note of
any interesting times, as you will want to explore these later using Stat!.
You will also want to make a note of the total number of deaths (on the screen following the S-shaped curve screen). We will use these to create a comparison of simulations, based on differences in the parameters of hierarchical versus contagious diffusion.
Use this web form to add your results to those of other students.
Here is the result file, if you want to check back later (you might be the first one to give some result!). Reload the file to see any updates.
You might try a few other choices of contagious versus hierarchical diffusion parameters, to augment our communal data file.
The software writes a file called diffuse.out in the directory containing the Diffuse executable. Be sure to save the file diffuse.out once you've chosen a simulation to explore further, however, as it is overwritten each time Diffuse runs.
We will now turn this output file (diffuse.out) into a set of files which can be used in either Stat!, GeoMed, or Gamma software (although we will only use the Stat! and GeoMed files). You will need to upload the file diffuse.out to a web-based converter: it will produce zip files containing the input files you need. Save the Stat! and GeoMed zip files to your computer.
Use the web-based converter now.
- We'll use the output files produced for Stat! to compare the two types
of diffusion you specified in your simulation model (remember that you
chose, and made note, of the two diffusion parameters - contagious versus
hierarchical).
- UMich Students: First download Stat! - ask! - and install it to the c:\temp\stat directory.
- You need to extract the Stat! files contained in the zip file, produced
by the web converter, to the same directory containing the Stat! executable
(Stat! is finicky that way).
- Now explore the Moran's I for the pnt files (diff0.pnt, etc.) using the
two different contiguity matrices:
- diffrook.con, which is the contiguity relationship based on Rook's contiguity (1 if the two locations are next to each other either horizontally or vertically; 0 otherwise); and
- diffhier.con, which is the contiguity relationship where a 1 indicates that two points are similar in population (i.e. either the next size up, the next size down, or the same size), and 0 otherwise.
Choose pnt files based on the S-shaped curve for your particular data set: at what timestep was the epidemic taking off? At what timestep was it leveling off?
Do you discover that there is a radical difference for some pnt files between the results obtained for the Moran's I using the two different types of contiguity matrices? Do the results jive with the parameters you chose for the different types of diffusion (i.e., if you selected a small contagious diffusion component and a large hierarchical coefficient, is this reflected in the behavior of Moran's I depending on contiguity matrix?
- UMich Students: First download Stat! - ask! - and install it to the c:\temp\stat directory.
- Now let's use GeoMed and Kulldorff's Spatial Scan statistic to
investigate space-time clusters in your simulated data set.
UMich Students: First download, and install GeoMed to the c:\temp directory.
- Extract the GeoMed files contained in the appropriate zip file produced by the web converter.
- Fire up GeoMed, and run the Space-time test (Kulldorff's Scan). If you use 99 simulations, this will take about 5 minutes to run!
- Note that the map is flipped upside down along the y-direction: while
Dr. Griffith's software plots the point (1,1) in the upper left corner,
GeoMed plots it in the lower left (Dr. Griffith plots his population map as
a matrix, while GeoMed plots it as geographical coordinates). So in this map,
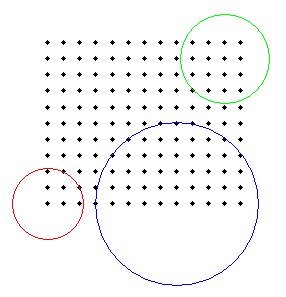
The lower-left and upper-right corners correspond to the most populated areas in his clustered population distribution. - How can you interpret the location (in space and time) of the three clusters? Do they make sense to you, based on your simulation model?
- Don't forget to evaluate the lab.
Principal readings:
- Smallman-Raynor, Matthew and Andrew D. Cliff. The Philippines insurrection and the 1902-4 cholera epidemic: Part I - Epidemiological diffusion processes in war Journal of Historical Geography, Vol. 24, No. 1. January 1998, pp. 69-89.
- Smallman-Raynor, Matthew and Andrew D. Cliff. The Philippines insurrection and the 1902-4 cholera epidemic: Part II - Diffusion Patterns in war and peace Journal of Historical Geography, Vol. 24, No. 1. January 1998, pp. 69-89.
- Gould, Peter. Spatial diffusion. Association of American Geographers, 1969. Series: Resource paper (Association of American Geographers. Commission on College Geography); no. 4. Especially pp. 1-23; 26-38 discuss the MIF (mean information field)
- Cliff, A.D. et al. Spatial diffusion : an historical geography of epidemics in an island community. Cambridge University Press, 1981.
- Gould, Peter. 1993, The Slow Plague, Blackwell; out of print.
- For information on the Christaller distribution: "10 Geographic Ideas that Changed the World," S. Hanson (ed). Rutgers U. Press, 1997.
- Lab Manual, in Word and html formats.
- Dr. Dan Griffith
- AIDS Data Animation Project
- FluTrack