Skip to main content\(\newenvironment{mat}{\left[\begin{array}}{\end{array}\right]}
\newcommand{\colvec}[1]{\left[\begin{matrix}#1 \end{matrix}\right]}
\newcommand{\rowvec}[1]{[\begin{matrix} #1 \end{matrix}]}
\newcommand{\definiteintegral}[4]{\int_{#1}^{#2}\,#3\,d#4}
\newcommand{\indefiniteintegral}[2]{\int#1\,d#2}
\def\u{\underline}
\def\summ{\sum\limits}
\newcommand{\lt}{ < }
\newcommand{\gt}{ > }
\newcommand{\amp}{ & }
\)
Section12SIR Model and Differential Equations¶ permalink
A simple model of infectious disease can be created in a visual and,
dare-I-say, even enjoyable way using a "free" website called
InsightMaker. These
"compartmental" models allow one to build models visually, by creating
"stocks" and "flows" and parameters, etc., and getting them connected
up the way you want. Then you can "simulate" the dynamics, illustrating
the facets you want with time-series, or scatterplots, or
what-have-you.
Anyway, this is all the commercial message you're going to get, but
it's a nice piece of software that gently introduces one to modeling
dynamical systems.
They also have a lot of nice demos on hand, created by individuals
willing to share, so I was able to pick up an SIR model ("clone" it)
from another user, and we've been playing with that.
So, once we're done building the model in InsightMaker, what have we
actually done? We've built a system of differential equations, which we
can solve using any standard mathematical software (e.g. Mathematica).
I want to take a look at the SIR from the differential equation
standpoint now, and reduce the system we've been studying down to its
sleekest, most elegant form. We do so, however, at the cost of some
mathematical analysis.
In the end, once all the flows have been set out, the parameters
described and their ranges set, we watch as the populations in the
"stocks" go through their motions. And we wonder "what kinds of dynamic
behaviors are possible?"
The visual elements of InsightMaker are translated into equations,
called differential equations, that are then solved
(numerically) by turning them into difference equations
(discrete versions of the continuous differential equations).
The flows that we've established, represented in this diagram,
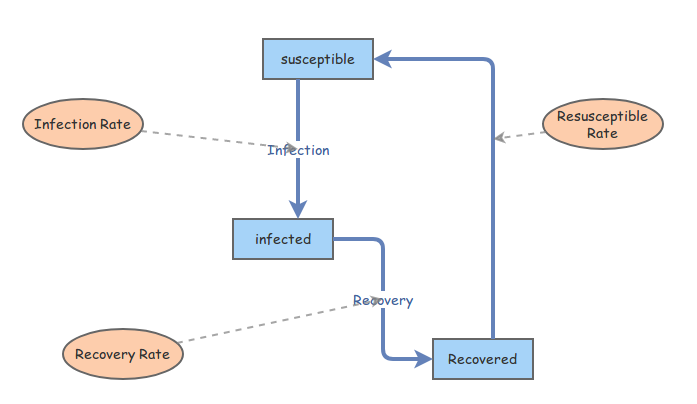 are converted into the following system of equations, which represent
the populations at the next moment, a timestep \(\Delta t\) later:
\begin{equation*}
\begin{array}{l}
\displaystyle
S(t+\Delta t) = S(t) + (-\alpha S(t)I(t) + \gamma R(t))\Delta t \cr
I(t+\Delta t) = I(t) + ( \alpha S(t)I(t) - \beta I(t))\Delta t \cr
R(t+\Delta t) = R(t) + (-\gamma R(t) + \beta I(t))\Delta t
\end{array}
\end{equation*}
where \(\alpha\) is the infection "rate", \(\beta\) is the
recovery rate, and \(\gamma\) is the resusceptible rate. Notice that
I put the infectivity "rate" in quotes: there's an issue that we're
going to discuss in a moment.
Now divide each side by \(\Delta t\), and we've got
\begin{equation*}
\begin{array}{l}
\displaystyle
\frac{S(t+\Delta t)-S(t)}{\Delta t} =-\alpha S(t)I(t) + \gamma R(t) \cr
\frac{I(t+\Delta t)-I(t)}{\Delta t} = \alpha S(t)I(t) - \beta I(t) \cr
\frac{R(t+\Delta t)-R(t)}{\Delta t} = -\gamma R(t) + \beta I(t)
\end{array}
\end{equation*}
Then take a limit as \(\Delta t \to 0\), and we finally have our
system of differential equations:
\begin{equation*}
\begin{array}{l}
\displaystyle
S'(t) =-\alpha S(t)I(t) + \gamma R(t) \cr
I'(t) = \alpha S(t)I(t) - \beta I(t) \cr
R'(t) = -\gamma R(t) + \beta I(t)
\end{array}
\end{equation*}
Okay: if you scaled the sub-populations by multiplying each by 100, should
that change the dynamics? Or would everything proceed the same, just on
a 100-fold larger scale? I would think that everything would look the
same, just on a 100-fold larger scale. However, if you scale each
sub-population by 100, the dynamics change. That's because of the
"bi-linear" term \(S(t)I(t)\): we would have to divide \(\alpha\)
by 100 to keep the same dynamics. That's a problem, and one which shows
that \(\alpha\) is not the rate that we expect. I'll handle that
problem in a moment.
First, let's consider the total
population, \(N(t)=S(t)+I(t)+R(t)\). It
turns out that \(N(t)\) is actually constant, since
\begin{equation*}
\frac{dN(t)}{dt}=\frac{d}{dt}(S(t)+I(t)+R(t))=0
\end{equation*}
(just add all the right-hand sides of the system of equations to see
that they cancel each other).
This just says that no one is lost to the system - they stay in one
population or the other, and no people are created or
destroyed. \(N(t)\) called a conserved
quantity. Since \(N(t)\) is actually constant, let's just call
it \(N\).
If we divide all the populations by N, then they're turned into
proportions, which sum to 1.
Now back to the problem mentioned just a moment ago. Actually, the
interaction term, which models the infection flow, has the wrong units
if we think of \(\alpha\) as a rate (the rates have units
of \(\frac{1}{time}\): however, if we change that flow from
\begin{equation*}\alpha S(t)I(t)\end{equation*}
to
\begin{equation*}\alpha S(t)\frac{I(t)}{N}\end{equation*}
then we're okay.
It also changes how we think about the interaction. We
still have a bi-linear relationship: if you double the infected
population, you'll double the number of infected susceptibles; and if
you double the number of susceptibles, you'll double the number of
infected susceptibles. But it now has the units
of \(\frac{people}{time}\), which is then multiplied by \(\Delta
t\) to turn that flow into a flow of \(people\)!
Think of \(\frac{I(t)}{N}\) as the proportional to the probability
that a susceptible will bump into an infected person.
Okay: now we've got a new system, where every sub-population has been scaled
by dividing by \(N\):
\begin{equation*}
\begin{array}{l}
\displaystyle
s'(t) =-\alpha s(t)i(t) + \gamma r(t) \cr
i'(t) = \alpha s(t)i(t) - \beta i(t) \cr
r'(t) = -\gamma r(t) + \beta i(t)
\end{array}
\end{equation*}
Now, for our final trick, we divide each side of each equation by \(\alpha\):
\begin{equation*}
\begin{array}{l}
\displaystyle
\frac{ds}{\alpha dt} = -s(t)i(t) + \frac{\gamma}{\alpha} r(t) \cr
\frac{di}{\alpha dt} = s(t)i(t) - \frac{\beta}{\alpha} i(t) \cr
\frac{dr}{\alpha dt} = -\frac{\gamma}{\alpha} r(t) + \frac{\beta}{\alpha} i(t)
\end{array}
\end{equation*}
and we define new parameters
\begin{equation*}
\begin{array}{l}
\displaystyle
\frac{ds}{\alpha dt} = -s(t)i(t) + \mu r(t) \cr
\frac{di}{\alpha dt} = s(t)i(t) - \lambda i(t) \cr
\frac{dr}{\alpha dt} = -\mu r(t) + \lambda i(t)
\end{array}
\end{equation*}
where \(\lambda=\frac{\beta}{\alpha}\),
and \(\mu=\frac{\gamma}{\alpha}\). These are relative rates,
rates relative to \(\alpha\), and the only two parameters left in
our model. As we play with these parameters, we'll see all the dynamics
possible in this system.
If we now rescale time, as \(\tau=\alpha t\); then, by the chain rule,
\begin{equation*}\frac{ds}{\alpha dt}=\frac{ds}{\alpha
d\tau}\frac{d\tau}{dt}=\frac{ds}{\alpha d\tau}\alpha=\frac{ds}{d\tau}\end{equation*}
What does this rescaling of time mean? Let's consider an example, where
the infection rate is \(\alpha=1/100/day\). Then if \(\tau=1\),
then \(\alpha t=1\), so \(t=100\) days. So time is relative to
the disease's infection rate.
What this tells us is that the qualitative picture of two diseases with
a different values of \(\alpha\) will look qualitatively the same
dynamically, provided the other rate ratios are equal.
So the simplest, sleekest equations are these:
\begin{equation*}
\begin{array}{l}
\displaystyle
\frac{ds}{d\tau} = -s(\tau)i(\tau) + \mu r(\tau) \cr
\frac{di}{d\tau} = s(\tau)i(\tau) - \lambda i(\tau) \cr
\frac{dr}{d\tau} = -\mu r(\tau) + \lambda i(\tau)
\end{array}
\end{equation*}
In the end, there's not a lot that can happen: either the infection
dies out, or it becomes endemic, and we reach a "steady state", wherein
each of the three populations tends towards an asymptotically stable
value.
To find the steady state, you need to find a set \((s^*,i^*,r^*)\)
which makes each right-hand side of the system of equations above
exactly zero - which means that the sub-populations are not changing in
time (their derivatives are zero). That's called a fixed point
of the system. Then the question becomes "Is it stable?": that is, if
we push the system away from the fixed point, does it come back, or fly
away?
The parameter \(\lambda\) is critical. It turns out that
if \(\lambda\) (which is the ratio of recovery-to-infection rates)
is greater than 1, then the infection will die out; if it is less than
1, then the infection will become endemic - remain forever in the
population. So it's really important.
are converted into the following system of equations, which represent
the populations at the next moment, a timestep \(\Delta t\) later:
\begin{equation*}
\begin{array}{l}
\displaystyle
S(t+\Delta t) = S(t) + (-\alpha S(t)I(t) + \gamma R(t))\Delta t \cr
I(t+\Delta t) = I(t) + ( \alpha S(t)I(t) - \beta I(t))\Delta t \cr
R(t+\Delta t) = R(t) + (-\gamma R(t) + \beta I(t))\Delta t
\end{array}
\end{equation*}
where \(\alpha\) is the infection "rate", \(\beta\) is the
recovery rate, and \(\gamma\) is the resusceptible rate. Notice that
I put the infectivity "rate" in quotes: there's an issue that we're
going to discuss in a moment.
Now divide each side by \(\Delta t\), and we've got
\begin{equation*}
\begin{array}{l}
\displaystyle
\frac{S(t+\Delta t)-S(t)}{\Delta t} =-\alpha S(t)I(t) + \gamma R(t) \cr
\frac{I(t+\Delta t)-I(t)}{\Delta t} = \alpha S(t)I(t) - \beta I(t) \cr
\frac{R(t+\Delta t)-R(t)}{\Delta t} = -\gamma R(t) + \beta I(t)
\end{array}
\end{equation*}
Then take a limit as \(\Delta t \to 0\), and we finally have our
system of differential equations:
\begin{equation*}
\begin{array}{l}
\displaystyle
S'(t) =-\alpha S(t)I(t) + \gamma R(t) \cr
I'(t) = \alpha S(t)I(t) - \beta I(t) \cr
R'(t) = -\gamma R(t) + \beta I(t)
\end{array}
\end{equation*}
Okay: if you scaled the sub-populations by multiplying each by 100, should
that change the dynamics? Or would everything proceed the same, just on
a 100-fold larger scale? I would think that everything would look the
same, just on a 100-fold larger scale. However, if you scale each
sub-population by 100, the dynamics change. That's because of the
"bi-linear" term \(S(t)I(t)\): we would have to divide \(\alpha\)
by 100 to keep the same dynamics. That's a problem, and one which shows
that \(\alpha\) is not the rate that we expect. I'll handle that
problem in a moment.
First, let's consider the total
population, \(N(t)=S(t)+I(t)+R(t)\). It
turns out that \(N(t)\) is actually constant, since
\begin{equation*}
\frac{dN(t)}{dt}=\frac{d}{dt}(S(t)+I(t)+R(t))=0
\end{equation*}
(just add all the right-hand sides of the system of equations to see
that they cancel each other).
This just says that no one is lost to the system - they stay in one
population or the other, and no people are created or
destroyed. \(N(t)\) called a conserved
quantity. Since \(N(t)\) is actually constant, let's just call
it \(N\).
If we divide all the populations by N, then they're turned into
proportions, which sum to 1.
Now back to the problem mentioned just a moment ago. Actually, the
interaction term, which models the infection flow, has the wrong units
if we think of \(\alpha\) as a rate (the rates have units
of \(\frac{1}{time}\): however, if we change that flow from
\begin{equation*}\alpha S(t)I(t)\end{equation*}
to
\begin{equation*}\alpha S(t)\frac{I(t)}{N}\end{equation*}
then we're okay.
It also changes how we think about the interaction. We
still have a bi-linear relationship: if you double the infected
population, you'll double the number of infected susceptibles; and if
you double the number of susceptibles, you'll double the number of
infected susceptibles. But it now has the units
of \(\frac{people}{time}\), which is then multiplied by \(\Delta
t\) to turn that flow into a flow of \(people\)!
Think of \(\frac{I(t)}{N}\) as the proportional to the probability
that a susceptible will bump into an infected person.
Okay: now we've got a new system, where every sub-population has been scaled
by dividing by \(N\):
\begin{equation*}
\begin{array}{l}
\displaystyle
s'(t) =-\alpha s(t)i(t) + \gamma r(t) \cr
i'(t) = \alpha s(t)i(t) - \beta i(t) \cr
r'(t) = -\gamma r(t) + \beta i(t)
\end{array}
\end{equation*}
Now, for our final trick, we divide each side of each equation by \(\alpha\):
\begin{equation*}
\begin{array}{l}
\displaystyle
\frac{ds}{\alpha dt} = -s(t)i(t) + \frac{\gamma}{\alpha} r(t) \cr
\frac{di}{\alpha dt} = s(t)i(t) - \frac{\beta}{\alpha} i(t) \cr
\frac{dr}{\alpha dt} = -\frac{\gamma}{\alpha} r(t) + \frac{\beta}{\alpha} i(t)
\end{array}
\end{equation*}
and we define new parameters
\begin{equation*}
\begin{array}{l}
\displaystyle
\frac{ds}{\alpha dt} = -s(t)i(t) + \mu r(t) \cr
\frac{di}{\alpha dt} = s(t)i(t) - \lambda i(t) \cr
\frac{dr}{\alpha dt} = -\mu r(t) + \lambda i(t)
\end{array}
\end{equation*}
where \(\lambda=\frac{\beta}{\alpha}\),
and \(\mu=\frac{\gamma}{\alpha}\). These are relative rates,
rates relative to \(\alpha\), and the only two parameters left in
our model. As we play with these parameters, we'll see all the dynamics
possible in this system.
If we now rescale time, as \(\tau=\alpha t\); then, by the chain rule,
\begin{equation*}\frac{ds}{\alpha dt}=\frac{ds}{\alpha
d\tau}\frac{d\tau}{dt}=\frac{ds}{\alpha d\tau}\alpha=\frac{ds}{d\tau}\end{equation*}
What does this rescaling of time mean? Let's consider an example, where
the infection rate is \(\alpha=1/100/day\). Then if \(\tau=1\),
then \(\alpha t=1\), so \(t=100\) days. So time is relative to
the disease's infection rate.
What this tells us is that the qualitative picture of two diseases with
a different values of \(\alpha\) will look qualitatively the same
dynamically, provided the other rate ratios are equal.
So the simplest, sleekest equations are these:
\begin{equation*}
\begin{array}{l}
\displaystyle
\frac{ds}{d\tau} = -s(\tau)i(\tau) + \mu r(\tau) \cr
\frac{di}{d\tau} = s(\tau)i(\tau) - \lambda i(\tau) \cr
\frac{dr}{d\tau} = -\mu r(\tau) + \lambda i(\tau)
\end{array}
\end{equation*}
In the end, there's not a lot that can happen: either the infection
dies out, or it becomes endemic, and we reach a "steady state", wherein
each of the three populations tends towards an asymptotically stable
value.
To find the steady state, you need to find a set \((s^*,i^*,r^*)\)
which makes each right-hand side of the system of equations above
exactly zero - which means that the sub-populations are not changing in
time (their derivatives are zero). That's called a fixed point
of the system. Then the question becomes "Is it stable?": that is, if
we push the system away from the fixed point, does it come back, or fly
away?
The parameter \(\lambda\) is critical. It turns out that
if \(\lambda\) (which is the ratio of recovery-to-infection rates)
is greater than 1, then the infection will die out; if it is less than
1, then the infection will become endemic - remain forever in the
population. So it's really important.
 are converted into the following system of equations, which represent
the populations at the next moment, a timestep \(\Delta t\) later:
\begin{equation*}
\begin{array}{l}
\displaystyle
S(t+\Delta t) = S(t) + (-\alpha S(t)I(t) + \gamma R(t))\Delta t \cr
I(t+\Delta t) = I(t) + ( \alpha S(t)I(t) - \beta I(t))\Delta t \cr
R(t+\Delta t) = R(t) + (-\gamma R(t) + \beta I(t))\Delta t
\end{array}
\end{equation*}
where \(\alpha\) is the infection "rate", \(\beta\) is the
recovery rate, and \(\gamma\) is the resusceptible rate. Notice that
I put the infectivity "rate" in quotes: there's an issue that we're
going to discuss in a moment.
Now divide each side by \(\Delta t\), and we've got
\begin{equation*}
\begin{array}{l}
\displaystyle
\frac{S(t+\Delta t)-S(t)}{\Delta t} =-\alpha S(t)I(t) + \gamma R(t) \cr
\frac{I(t+\Delta t)-I(t)}{\Delta t} = \alpha S(t)I(t) - \beta I(t) \cr
\frac{R(t+\Delta t)-R(t)}{\Delta t} = -\gamma R(t) + \beta I(t)
\end{array}
\end{equation*}
Then take a limit as \(\Delta t \to 0\), and we finally have our
system of differential equations:
\begin{equation*}
\begin{array}{l}
\displaystyle
S'(t) =-\alpha S(t)I(t) + \gamma R(t) \cr
I'(t) = \alpha S(t)I(t) - \beta I(t) \cr
R'(t) = -\gamma R(t) + \beta I(t)
\end{array}
\end{equation*}
Okay: if you scaled the sub-populations by multiplying each by 100, should
that change the dynamics? Or would everything proceed the same, just on
a 100-fold larger scale? I would think that everything would look the
same, just on a 100-fold larger scale. However, if you scale each
sub-population by 100, the dynamics change. That's because of the
"bi-linear" term \(S(t)I(t)\): we would have to divide \(\alpha\)
by 100 to keep the same dynamics. That's a problem, and one which shows
that \(\alpha\) is not the rate that we expect. I'll handle that
problem in a moment.
First, let's consider the total
population, \(N(t)=S(t)+I(t)+R(t)\). It
turns out that \(N(t)\) is actually constant, since
\begin{equation*}
\frac{dN(t)}{dt}=\frac{d}{dt}(S(t)+I(t)+R(t))=0
\end{equation*}
(just add all the right-hand sides of the system of equations to see
that they cancel each other).
This just says that no one is lost to the system - they stay in one
population or the other, and no people are created or
destroyed. \(N(t)\) called a conserved
quantity. Since \(N(t)\) is actually constant, let's just call
it \(N\).
If we divide all the populations by N, then they're turned into
proportions, which sum to 1.
Now back to the problem mentioned just a moment ago. Actually, the
interaction term, which models the infection flow, has the wrong units
if we think of \(\alpha\) as a rate (the rates have units
of \(\frac{1}{time}\): however, if we change that flow from
\begin{equation*}\alpha S(t)I(t)\end{equation*}
to
\begin{equation*}\alpha S(t)\frac{I(t)}{N}\end{equation*}
then we're okay.
It also changes how we think about the interaction. We
still have a bi-linear relationship: if you double the infected
population, you'll double the number of infected susceptibles; and if
you double the number of susceptibles, you'll double the number of
infected susceptibles. But it now has the units
of \(\frac{people}{time}\), which is then multiplied by \(\Delta
t\) to turn that flow into a flow of \(people\)!
Think of \(\frac{I(t)}{N}\) as the proportional to the probability
that a susceptible will bump into an infected person.
Okay: now we've got a new system, where every sub-population has been scaled
by dividing by \(N\):
\begin{equation*}
\begin{array}{l}
\displaystyle
s'(t) =-\alpha s(t)i(t) + \gamma r(t) \cr
i'(t) = \alpha s(t)i(t) - \beta i(t) \cr
r'(t) = -\gamma r(t) + \beta i(t)
\end{array}
\end{equation*}
Now, for our final trick, we divide each side of each equation by \(\alpha\):
\begin{equation*}
\begin{array}{l}
\displaystyle
\frac{ds}{\alpha dt} = -s(t)i(t) + \frac{\gamma}{\alpha} r(t) \cr
\frac{di}{\alpha dt} = s(t)i(t) - \frac{\beta}{\alpha} i(t) \cr
\frac{dr}{\alpha dt} = -\frac{\gamma}{\alpha} r(t) + \frac{\beta}{\alpha} i(t)
\end{array}
\end{equation*}
and we define new parameters
\begin{equation*}
\begin{array}{l}
\displaystyle
\frac{ds}{\alpha dt} = -s(t)i(t) + \mu r(t) \cr
\frac{di}{\alpha dt} = s(t)i(t) - \lambda i(t) \cr
\frac{dr}{\alpha dt} = -\mu r(t) + \lambda i(t)
\end{array}
\end{equation*}
where \(\lambda=\frac{\beta}{\alpha}\),
and \(\mu=\frac{\gamma}{\alpha}\). These are relative rates,
rates relative to \(\alpha\), and the only two parameters left in
our model. As we play with these parameters, we'll see all the dynamics
possible in this system.
If we now rescale time, as \(\tau=\alpha t\); then, by the chain rule,
\begin{equation*}\frac{ds}{\alpha dt}=\frac{ds}{\alpha
d\tau}\frac{d\tau}{dt}=\frac{ds}{\alpha d\tau}\alpha=\frac{ds}{d\tau}\end{equation*}
What does this rescaling of time mean? Let's consider an example, where
the infection rate is \(\alpha=1/100/day\). Then if \(\tau=1\),
then \(\alpha t=1\), so \(t=100\) days. So time is relative to
the disease's infection rate.
What this tells us is that the qualitative picture of two diseases with
a different values of \(\alpha\) will look qualitatively the same
dynamically, provided the other rate ratios are equal.
So the simplest, sleekest equations are these:
\begin{equation*}
\begin{array}{l}
\displaystyle
\frac{ds}{d\tau} = -s(\tau)i(\tau) + \mu r(\tau) \cr
\frac{di}{d\tau} = s(\tau)i(\tau) - \lambda i(\tau) \cr
\frac{dr}{d\tau} = -\mu r(\tau) + \lambda i(\tau)
\end{array}
\end{equation*}
In the end, there's not a lot that can happen: either the infection
dies out, or it becomes endemic, and we reach a "steady state", wherein
each of the three populations tends towards an asymptotically stable
value.
To find the steady state, you need to find a set \((s^*,i^*,r^*)\)
which makes each right-hand side of the system of equations above
exactly zero - which means that the sub-populations are not changing in
time (their derivatives are zero). That's called a fixed point
of the system. Then the question becomes "Is it stable?": that is, if
we push the system away from the fixed point, does it come back, or fly
away?
The parameter \(\lambda\) is critical. It turns out that
if \(\lambda\) (which is the ratio of recovery-to-infection rates)
is greater than 1, then the infection will die out; if it is less than
1, then the infection will become endemic - remain forever in the
population. So it's really important.
are converted into the following system of equations, which represent
the populations at the next moment, a timestep \(\Delta t\) later:
\begin{equation*}
\begin{array}{l}
\displaystyle
S(t+\Delta t) = S(t) + (-\alpha S(t)I(t) + \gamma R(t))\Delta t \cr
I(t+\Delta t) = I(t) + ( \alpha S(t)I(t) - \beta I(t))\Delta t \cr
R(t+\Delta t) = R(t) + (-\gamma R(t) + \beta I(t))\Delta t
\end{array}
\end{equation*}
where \(\alpha\) is the infection "rate", \(\beta\) is the
recovery rate, and \(\gamma\) is the resusceptible rate. Notice that
I put the infectivity "rate" in quotes: there's an issue that we're
going to discuss in a moment.
Now divide each side by \(\Delta t\), and we've got
\begin{equation*}
\begin{array}{l}
\displaystyle
\frac{S(t+\Delta t)-S(t)}{\Delta t} =-\alpha S(t)I(t) + \gamma R(t) \cr
\frac{I(t+\Delta t)-I(t)}{\Delta t} = \alpha S(t)I(t) - \beta I(t) \cr
\frac{R(t+\Delta t)-R(t)}{\Delta t} = -\gamma R(t) + \beta I(t)
\end{array}
\end{equation*}
Then take a limit as \(\Delta t \to 0\), and we finally have our
system of differential equations:
\begin{equation*}
\begin{array}{l}
\displaystyle
S'(t) =-\alpha S(t)I(t) + \gamma R(t) \cr
I'(t) = \alpha S(t)I(t) - \beta I(t) \cr
R'(t) = -\gamma R(t) + \beta I(t)
\end{array}
\end{equation*}
Okay: if you scaled the sub-populations by multiplying each by 100, should
that change the dynamics? Or would everything proceed the same, just on
a 100-fold larger scale? I would think that everything would look the
same, just on a 100-fold larger scale. However, if you scale each
sub-population by 100, the dynamics change. That's because of the
"bi-linear" term \(S(t)I(t)\): we would have to divide \(\alpha\)
by 100 to keep the same dynamics. That's a problem, and one which shows
that \(\alpha\) is not the rate that we expect. I'll handle that
problem in a moment.
First, let's consider the total
population, \(N(t)=S(t)+I(t)+R(t)\). It
turns out that \(N(t)\) is actually constant, since
\begin{equation*}
\frac{dN(t)}{dt}=\frac{d}{dt}(S(t)+I(t)+R(t))=0
\end{equation*}
(just add all the right-hand sides of the system of equations to see
that they cancel each other).
This just says that no one is lost to the system - they stay in one
population or the other, and no people are created or
destroyed. \(N(t)\) called a conserved
quantity. Since \(N(t)\) is actually constant, let's just call
it \(N\).
If we divide all the populations by N, then they're turned into
proportions, which sum to 1.
Now back to the problem mentioned just a moment ago. Actually, the
interaction term, which models the infection flow, has the wrong units
if we think of \(\alpha\) as a rate (the rates have units
of \(\frac{1}{time}\): however, if we change that flow from
\begin{equation*}\alpha S(t)I(t)\end{equation*}
to
\begin{equation*}\alpha S(t)\frac{I(t)}{N}\end{equation*}
then we're okay.
It also changes how we think about the interaction. We
still have a bi-linear relationship: if you double the infected
population, you'll double the number of infected susceptibles; and if
you double the number of susceptibles, you'll double the number of
infected susceptibles. But it now has the units
of \(\frac{people}{time}\), which is then multiplied by \(\Delta
t\) to turn that flow into a flow of \(people\)!
Think of \(\frac{I(t)}{N}\) as the proportional to the probability
that a susceptible will bump into an infected person.
Okay: now we've got a new system, where every sub-population has been scaled
by dividing by \(N\):
\begin{equation*}
\begin{array}{l}
\displaystyle
s'(t) =-\alpha s(t)i(t) + \gamma r(t) \cr
i'(t) = \alpha s(t)i(t) - \beta i(t) \cr
r'(t) = -\gamma r(t) + \beta i(t)
\end{array}
\end{equation*}
Now, for our final trick, we divide each side of each equation by \(\alpha\):
\begin{equation*}
\begin{array}{l}
\displaystyle
\frac{ds}{\alpha dt} = -s(t)i(t) + \frac{\gamma}{\alpha} r(t) \cr
\frac{di}{\alpha dt} = s(t)i(t) - \frac{\beta}{\alpha} i(t) \cr
\frac{dr}{\alpha dt} = -\frac{\gamma}{\alpha} r(t) + \frac{\beta}{\alpha} i(t)
\end{array}
\end{equation*}
and we define new parameters
\begin{equation*}
\begin{array}{l}
\displaystyle
\frac{ds}{\alpha dt} = -s(t)i(t) + \mu r(t) \cr
\frac{di}{\alpha dt} = s(t)i(t) - \lambda i(t) \cr
\frac{dr}{\alpha dt} = -\mu r(t) + \lambda i(t)
\end{array}
\end{equation*}
where \(\lambda=\frac{\beta}{\alpha}\),
and \(\mu=\frac{\gamma}{\alpha}\). These are relative rates,
rates relative to \(\alpha\), and the only two parameters left in
our model. As we play with these parameters, we'll see all the dynamics
possible in this system.
If we now rescale time, as \(\tau=\alpha t\); then, by the chain rule,
\begin{equation*}\frac{ds}{\alpha dt}=\frac{ds}{\alpha
d\tau}\frac{d\tau}{dt}=\frac{ds}{\alpha d\tau}\alpha=\frac{ds}{d\tau}\end{equation*}
What does this rescaling of time mean? Let's consider an example, where
the infection rate is \(\alpha=1/100/day\). Then if \(\tau=1\),
then \(\alpha t=1\), so \(t=100\) days. So time is relative to
the disease's infection rate.
What this tells us is that the qualitative picture of two diseases with
a different values of \(\alpha\) will look qualitatively the same
dynamically, provided the other rate ratios are equal.
So the simplest, sleekest equations are these:
\begin{equation*}
\begin{array}{l}
\displaystyle
\frac{ds}{d\tau} = -s(\tau)i(\tau) + \mu r(\tau) \cr
\frac{di}{d\tau} = s(\tau)i(\tau) - \lambda i(\tau) \cr
\frac{dr}{d\tau} = -\mu r(\tau) + \lambda i(\tau)
\end{array}
\end{equation*}
In the end, there's not a lot that can happen: either the infection
dies out, or it becomes endemic, and we reach a "steady state", wherein
each of the three populations tends towards an asymptotically stable
value.
To find the steady state, you need to find a set \((s^*,i^*,r^*)\)
which makes each right-hand side of the system of equations above
exactly zero - which means that the sub-populations are not changing in
time (their derivatives are zero). That's called a fixed point
of the system. Then the question becomes "Is it stable?": that is, if
we push the system away from the fixed point, does it come back, or fly
away?
The parameter \(\lambda\) is critical. It turns out that
if \(\lambda\) (which is the ratio of recovery-to-infection rates)
is greater than 1, then the infection will die out; if it is less than
1, then the infection will become endemic - remain forever in the
population. So it's really important.